Part:BBa_K1453002 Deleted
pBluescript II KS(+) ScaI deletion
Vector
We used pBluescript II KS(+) as our origin vector and then transformed it into the Connector of our desire.
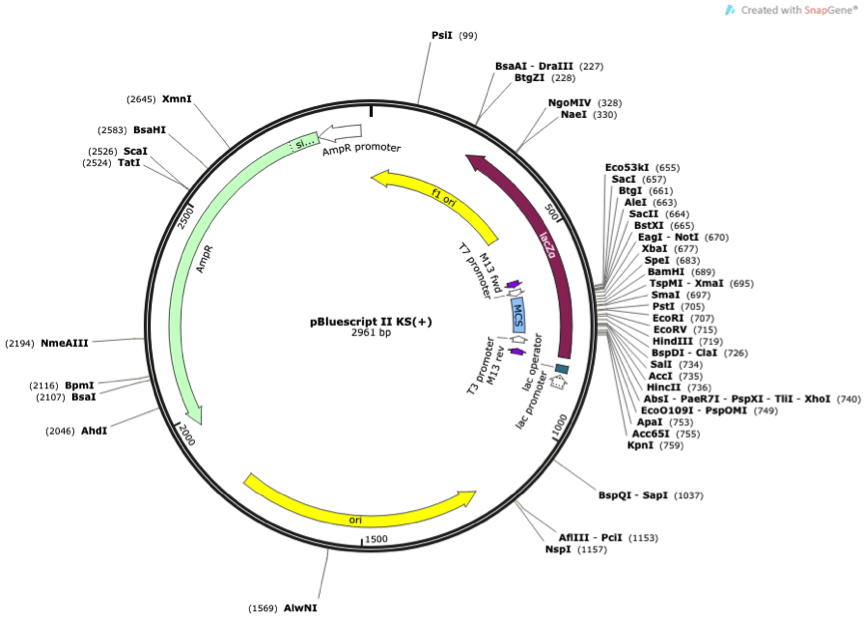
Figure 1 Vector map of pBluescript II KS(+)
Connector
The so-called Connector is plasmid designed to bind with several different Connectees(iGEM12_SJTU_BioX_Shanghai BBa_K771000). Connectees are fusion proteins who carry out functional enzymes and have TAL(Transactivator-like effectors) at one end that can bind to certain DNA sequences and you can design your own Connectees with different enzymes. For pBluescript II KS(+), we have selected three 14-nucleotide-long sequences called RSⅠ, RSⅡ and RSⅢ as the recognition sequences(RS). The recognition sequences must start with a T and end with a T. RS Ⅰ: TTCGATATCAAGCT RS Ⅱ: TGTGACTGGTGAGT RS Ⅲ: TTTGGTCATGAGAT ( RS Ⅰ contains partial restriction enzyme cutting site of EcoRI and EcoRV ) ( RS Ⅱ contains partial restriction enzyme cutting site of ScaI )
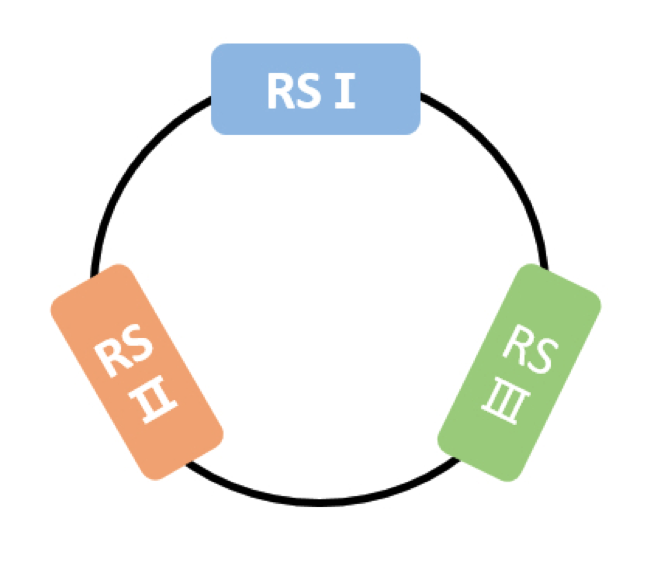 Figure 2 A Connector binds with three different Connectees
Figure 2 A Connector binds with three different Connectees
Combination
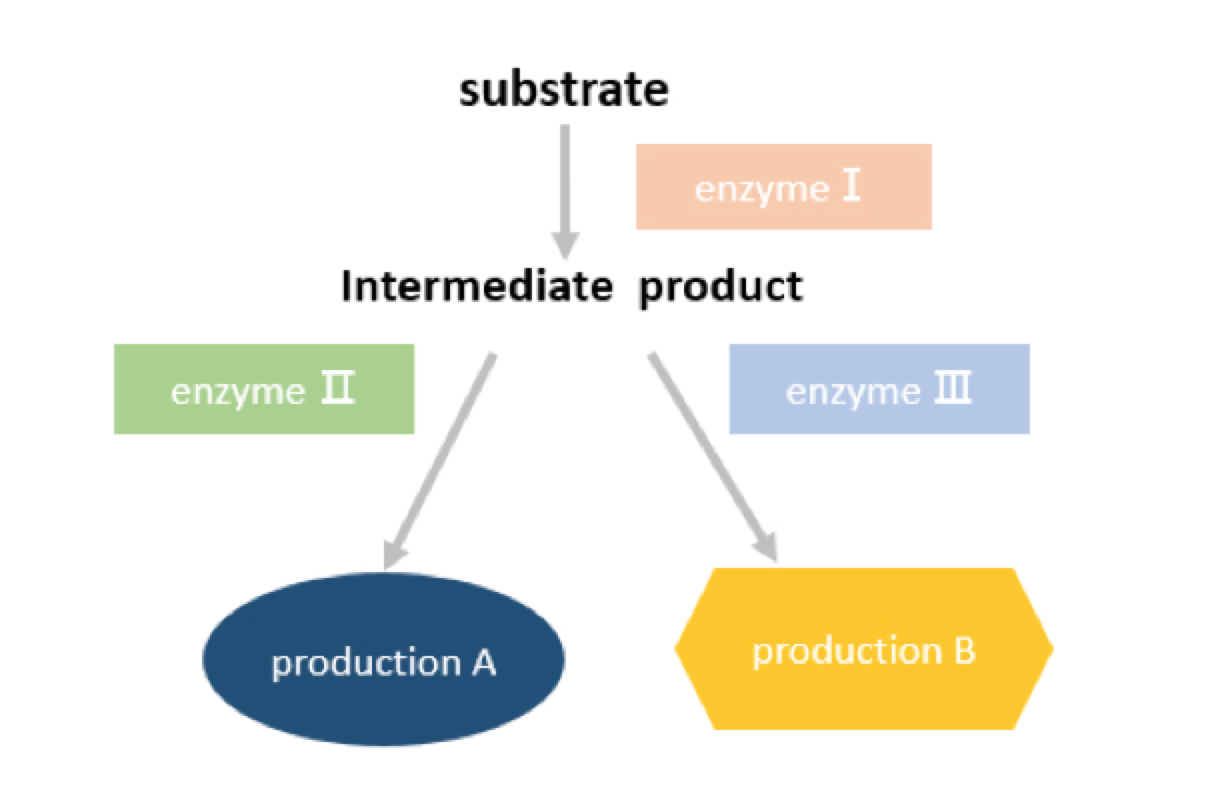
In order to prove that Connectors are able to bind with three kinds of Connectees. We designed the combination experiment. We selected a pathway: substrate becomes intermediate product through enzymeⅠ,then the intermediate product can either go to production A through enzymeⅡ or production B through enzyme Ⅲ.
Figure 3 Diagrams of our pathway design
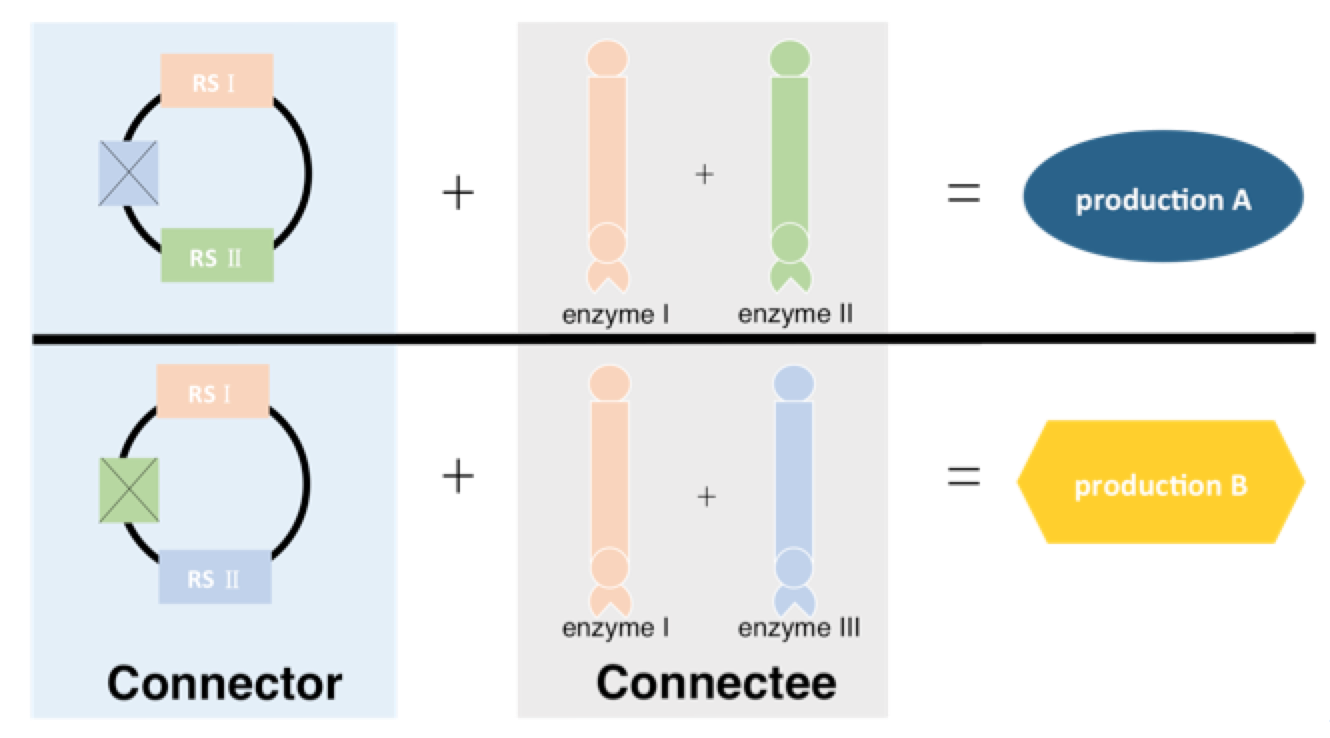
To achieve this, we need three kinds of Connectees fused with enzyme Ⅰ, ⅡandⅢ independently and two kinds of Connectors. One has RSⅠand RSⅡ while the other has RSⅠand RSⅢ. So in the end, Connectors with RSⅠ and RSⅡ get production A while Connectors with RSⅠ and RSⅢ production B.
Figure 4 Connector is originally designed with three recognition sequence(RS). Then we transformed it into two different Connector, one with RS Ⅰ and RS Ⅱ while the other one with RS Ⅰ and RS Ⅲ. Connector with RSⅠand RSⅡ can bind with Connectee-enzyme Ⅰ and Connectee-enzyme Ⅱ and get production A in the end, while Connector with RS Ⅰ and RS Ⅲ can bind with Connectee-enzyme Ⅰ and Connectee-enzyme Ⅲ and get production B. The corresponding Connectors are pBluescript II KS(+) ScaI deletion and pBluescript II KS(+) EcoRV deletion.
pBluescript II KS(+) ScaI deletion:
We delete the RSⅡ site so this transformed Connector can only bind to Connectees with two kinds of enzyme. In order to delete this site and surrounding sequence, we adopted the Inverse PCR to delete the following sequence: CTGTGACTGGTGAGTACTCAACCAAGTCATTCTG
Primers for SacⅠ: Forward: AGAATAGTGTATGCGGCGCGAC Tm:57℃ Reverse: AAAAGCATCTTACGGATGGCA Tm:58℃
pBluescript II KS(+) ScaI deletion can be used along with pBluescript IIKS(+) EcoRV deletion and our Connectees(iGEM12_SJTU_BioX_Shanghai BBa_K771000) to achieve your certain pathway design.
Inverse PCR
We used Inverse PCR to transform our plasmid into two kinds of Connectors: pBluescript II KS(+) ScaI deletion and pBluescript II KS(+) EcoRV deletion. Inverse PCR method is using cyclic DNA (such as a plasmid) as the template, with two primers designed in a reverse direction to achieve completed PCR. In this way, by designing primers,we can introduce a mutation, insertion or deletion. We used KOD-Plus-Mutagenesis Kit by TOYOBO.CO.LTD. More details please click http://www.bio-toyobo.cn.
Usage and Biology
If you want to use our Connector, there is a list for possible TAL recognition sequences on pBluescript II KS(+). You can choose any combination of following sequences to design your own Connectees.
ctaaattgtaagcgttaatattttgttaaaattcgcgttaaatttttgttaaatcagctcattttttaaccaataggccg aaatcggcaaaatcccttataaatcaaaagaatagaccgagatagggttgagtgttgttccagtttggaacaagagtcca ctattaaagaacgtggactccaacgtcaaagggcgaaaaaccgtctatcagggcgatggcccactacgtgaaccatcacc ctaatcaagttttttggggtcgaggtgccgtaaagcactaaatcggaaccctaaagggagcccccgatttagagcttgac ggggaaagccggcgaacgtggcgagaaaggaagggaagaaagcgaaaggagcgggcgctagggcgctggcaagtgtagcg gtcacgctgcgcgtaaccaccacacccgccgcgcttaatgcgccgctacagggcgcgtcccattcgccattcaggctgcg caactgttgggaagggcgatcggtgcgggcctcttcgctattacgccagctggcgaaagggggatgtgctgcaaggcgat taagttgggtaacgccagggttttcccagtcacgacgttgtaaaacgacggccagtgagcgcgcgtaatacgactcacta tagggcgaattggagctccaccgcggtggcggccgctctagaactagtggatcccccgggctgcaggaattcgatatcaa gcttatcgataccgtcgacctcgagggggggcccggtacccagcttttgttccctttagtgagggttaattgcgcgcttg gcgtaatcatggtcatagctgtttcctgtgtgaaattgttatccgctcacaattccacacaacatacgagccggaagcat aaagtgtaaagcctggggtgcctaatgagtgagctaactcacattaattgcgttgcgctcactgcccgctttccagtcgg gaaacctgtcgtgccagctgcattaatgaatcggccaacgcgcggggagaggcggtttgcgtattgggcgctcttccgct tcctcgctcactgactcgctgcgctcggtcgttcggctgcggcgagcggtatcagctcactcaaaggcggtaatacggtt atccacagaatcaggggataacgcaggaaagaacatgtgagcaaaaggccagcaaaaggccaggaaccgtaaaaaggccg cgttgctggcgtttttccataggctccgcccccctgacgagcatcacaaaaatcgacgctcaagtcagaggtggcgaaac ccgacaggactataaagataccaggcgtttccccctggaagctccctcgtgcgctctcctgttccgaccctgccgcttac cggatacctgtccgcctttctcccttcgggaagcgtggcgctttctcatagctcacgctgtaggtatctcagttcggtgt aggtcgttcgctccaagctgggctgtgtgcacgaaccccccgttcagcccgaccgctgcgccttatccggtaactatcgt cttgagtccaacccggtaagacacgacttatcgccactggcagcagccactggtaacaggattagcagagcgaggtatgt aggcggtgctacagagttcttgaagtggtggcctaactacggctacactagaaggacagtatttggtatctgcgctctgc tgaagccagttaccttcggaaaaagagttggtagctcttgatccggcaaacaaaccaccgctggtagcggtggttttttt gtttgcaagcagcagattacgcgcagaaaaaaaggatctcaagaagatcctttgatcttttctacggggtctgacgctca gtggaacgaaaactcacgttaagggattttggtcatgagattatcaaaaaggatcttcacctagatccttttaaattaaa aatgaagttttaaatcaatctaaagtatatatgagtaaacttggtctgacagttaccaatgcttaatcagtgaggcacct atctcagcgatctgtctatttcgttcatccatagttgcctgactccccgtcgtgtagataactacgatacgggagggctt accatctggccccagtgctgcaatgataccgcgagacccacgctcaccggctccagatttatcagcaataaaccagccag ccggaagggccgagcgcagaagtggtcctgcaactttatccgcctccatccagtctattaattgttgccgggaagctaga gtaagtagttcgccagttaatagtttgcgcaacgttgttgccattgctacaggcatcgtggtgtcacgctcgtcgtttgg tatggcttcattcagctccggttcccaacgatcaaggcgagttacatgatcccccatgttgtgcaaaaaagcggttagct ccttcggtcctccgatcgttgtcagaagtaagttggccgcagtgttatcactcatggttatggcagcactgcataattct cttactgtcatgccatccgtaagatgcttttagaatagtgtatgcggcgaccgagttgctcttgcccggcgtcaatacgg gataataccgcgccacatagcagaactttaaaagtgctcatcattggaaaacgttcttcggggcgaaaactctcaaggat cttaccgctgttgagatccagttcgatgtaacccactcgtgcacccaactgatcttcagcatcttttactttcaccagcg tttctgggtgagcaaaaacaggaaggcaaaatgccgcaaaaaagggaataagggcgacacggaaatgttgaatactcata ctcttcctttttcaatattattgaagcatttatcagggttattgtctcatgagcggatacatatttgaatgtatttagaa aaataaacaaataggggttccgcgcacatttccccgaaaagtgccac
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 707
Illegal XbaI site found at 677
Illegal SpeI site found at 683
Illegal PstI site found at 701 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 707
Illegal SpeI site found at 683
Illegal PstI site found at 701
Illegal NotI site found at 669 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 707
Illegal BamHI site found at 689
Illegal XhoI site found at 740 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 707
Illegal XbaI site found at 677
Illegal SpeI site found at 683
Illegal PstI site found at 701 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 707
Illegal XbaI site found at 677
Illegal SpeI site found at 683
Illegal PstI site found at 701
Illegal NgoMIV site found at 328 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 2113
Illegal SapI site found at 1030